Trajectory Preprocessing#
The following notebook illustrate the TrajPreprocessing class how to run the preprocessing steps.
Several preprocessing steps are available for the tail angle:
Interpolating missing values
Apply 1€ filter
The kinematic vigor is also computed from the speed and will also be useful for segmentation into bouts:
Loading dependencies
import numpy as np
import matplotlib.pyplot as plt
from megabouts.tracking_data import TrackingConfig, FullTrackingData, load_example_data
from megabouts.config import TrajPreprocessingConfig
from megabouts.preprocessing import TrajPreprocessing
Loading Data#
TrackingConfig and TrackingData similar to tutorial_Loading_Data
# Load data and set tracking configuration
df_recording, fps, mm_per_unit = load_example_data("SLEAP_fulltracking")
tracking_cfg = TrackingConfig(fps=fps, tracking="full_tracking")
# List of keypoints
keypoints = ["left_eye", "right_eye", "tail0", "tail1", "tail2", "tail3", "tail4"]
# Place NaN where the score is below a threshold
thresh_score = 0.0
for kps in keypoints:
score_below_thresh = df_recording["instance.score"] < thresh_score
df_recording.loc[
score_below_thresh | (df_recording[f"{kps}.score"] < thresh_score),
[f"{kps}.x", f"{kps}.y"],
] = np.nan
# Compute head and tail coordinates and convert to mm
head_x = ((df_recording["left_eye.x"] + df_recording["right_eye.x"]) / 2) * mm_per_unit
head_y = ((df_recording["left_eye.y"] + df_recording["right_eye.y"]) / 2) * mm_per_unit
tail_x = df_recording[[f"tail{i}.x" for i in range(5)]].values * mm_per_unit
tail_y = df_recording[[f"tail{i}.y" for i in range(5)]].values * mm_per_unit
# Create FullTrackingData object
tracking_data = FullTrackingData.from_keypoints(
head_x=head_x.values, head_y=head_y.values, tail_x=tail_x, tail_y=tail_y
)
Run Preprocessing#
Define preprocessing config
traj_preprocessing_cfg = TrajPreprocessingConfig(fps=tracking_cfg.fps)
Apply the trajectory preprocessing
traj_df_input = tracking_data.traj_df
traj = TrajPreprocessing(traj_preprocessing_cfg).preprocess_traj_df(traj_df_input)
traj.df contains information about the trajectory, the smooth values as well as the kinematic vigor:
traj.df.head(5)
| x | y | yaw | x_smooth | y_smooth | yaw_smooth | axial_speed | lateral_speed | yaw_speed | vigor | no_tracking | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 20.381254 | 20.996582 | 1.491111 | 20.381254 | 20.996582 | 1.491111 | NaN | NaN | NaN | 0.0 | 0.0 |
| 1 | 20.381340 | 20.996807 | 1.491013 | 20.381277 | 20.996642 | 1.491085 | NaN | NaN | NaN | 0.0 | 0.0 |
| 2 | 20.381337 | 20.997020 | 1.491086 | 20.381293 | 20.996742 | 1.491085 | NaN | NaN | NaN | 0.0 | 0.0 |
| 3 | 20.418551 | 20.997656 | 1.430916 | 20.391138 | 20.996983 | 1.475189 | NaN | NaN | NaN | 0.0 | 0.0 |
| 4 | 20.418671 | 20.997872 | 1.430749 | 20.399294 | 20.997219 | 1.461210 | 0.293531 | -1.616157 | -2.659344 | 0.0 | 0.0 |
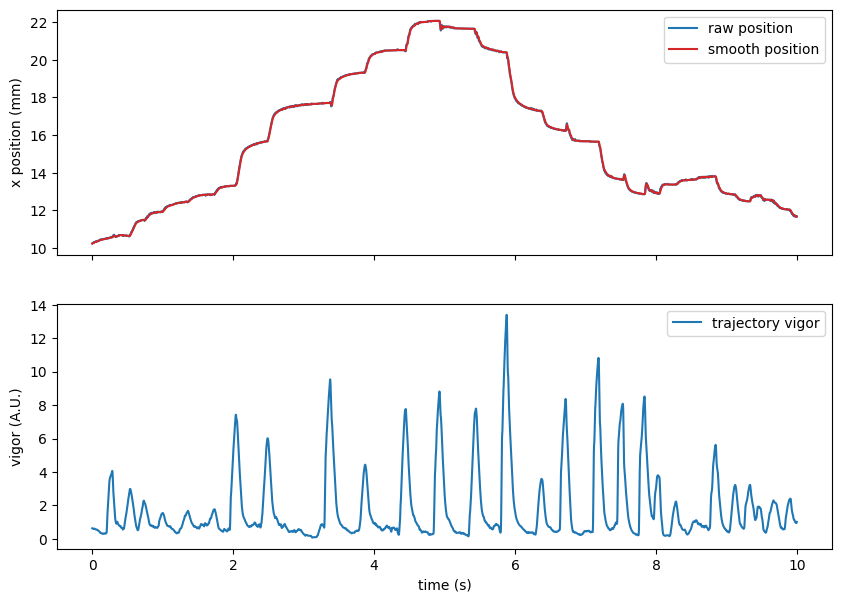
We can visualize the result of preprocessing:
Show code cell source
t = np.arange(tracking_data.T) / tracking_cfg.fps
IdSt = np.random.randint(tracking_data.T)
Duration = 10 * tracking_cfg.fps
t_win = t[IdSt : IdSt + Duration] - t[IdSt]
fig, ax = plt.subplots(2, 1, figsize=(10, 7), sharex=True)
ax[0].plot(t_win, traj.x[IdSt : IdSt + Duration], label="raw position")
ax[0].plot(
t_win,
traj.x_smooth[IdSt : IdSt + Duration],
label="smooth position",
color="tab:red",
)
ax[1].plot(t_win, traj.vigor[IdSt : IdSt + Duration], label="trajectory vigor")
ax[0].set(ylabel="x position (mm)")
ax[1].set(ylabel="vigor (A.U.)")
ax[1].set(xlabel="time (s)")
for i in range(2):
ax[i].legend()
plt.show()
Tuning 1€ filter#
📝 There are two configurable parameters in the filter, the minimum cutoff frequency
freq_cutoff_minand the speed coefficientbeta.
Decreasing the minimum cutoff frequency decreases slow speed jitter. Increasing the speed coefficient decreases speed lag.
The parameters are set using two-step procedure:
Step 1#
beta is set to 0 and freq_cutoff_min to 10Hz. Focus on a part of the recording where the fish is not swimming or swimming at low speed.
freq_cutoff_min is adjusted to remove jitter and preserve an acceptable lag during these slow movements.
traj_preprocessing_cfg = TrajPreprocessingConfig(
fps=tracking_cfg.fps, freq_cutoff_min=10, beta=0
)
Show code cell source
IdSt = 26800
Duration = 2 * tracking_cfg.fps
t_win = t[IdSt : IdSt + Duration] - t[IdSt]
x = traj_df_input.x
traj = TrajPreprocessing(traj_preprocessing_cfg).preprocess_traj_df(traj_df_input)
x_smooth = traj.df.x_smooth
fig, ax = plt.subplots(1, 1, figsize=(15, 4), sharex=True)
ax.set_title("Tuning " + r"$\text{freq_cutoff_min}$" + " during slow movement")
ax.plot(t_win, x[IdSt : IdSt + Duration], label="raw position")
ax.plot(
t_win, x_smooth[IdSt : IdSt + Duration], label="smoothed position", color="tab:red"
)
ax.set(xlabel="time (s)", ylabel="x position (mm)")
ax.legend()
plt.show()
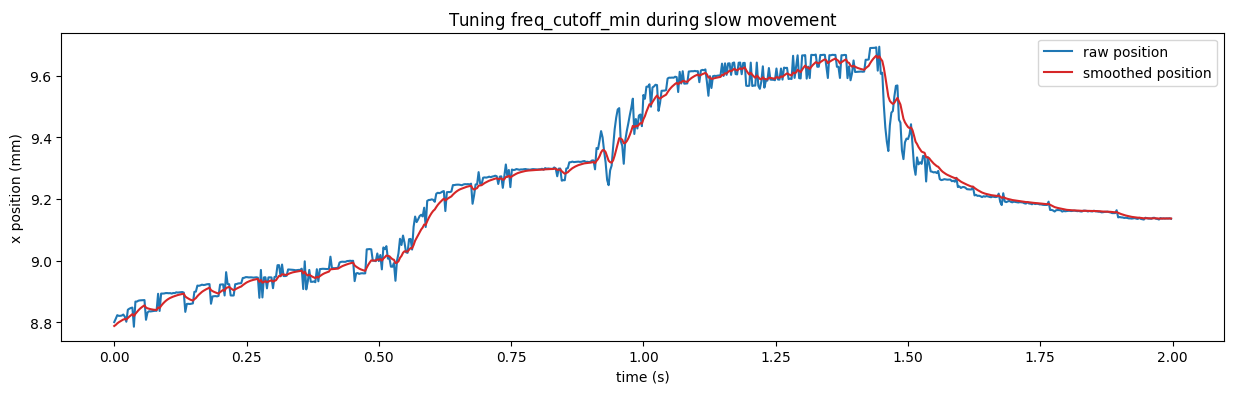
✅ Here a value close to 10Hz for freq_cutoff_min allows to smooth while keeping an acceptable lag
Step 2#
Focus on a part of the recording where the fish is moving fast. Adjust beta with a focus on minimizing lag.
traj_preprocessing_cfg = TrajPreprocessingConfig(
fps=tracking_cfg.fps, freq_cutoff_min=10, beta=1.4
)
Show code cell source
IdSt = 81600 # np.random.randint(x.shape[0])
Duration = 2 * tracking_cfg.fps
t_win = t[IdSt : IdSt + Duration] - t[IdSt]
x = traj_df_input.x
traj = TrajPreprocessing(traj_preprocessing_cfg).preprocess_traj_df(traj_df_input)
x_smooth = traj.df.x_smooth
fig, ax = plt.subplots(1, 1, figsize=(15, 4), sharex=True)
ax.set_title("Tuning " + r"$\text{beta}$" + " during fast movement")
ax.plot(t_win, x[IdSt : IdSt + Duration], label="raw position")
ax.plot(
t_win, x_smooth[IdSt : IdSt + Duration], label="smoothed position", color="tab:red"
)
ax.set(xlabel="time (s)", ylabel="x position (mm)")
ax.legend()
plt.show()
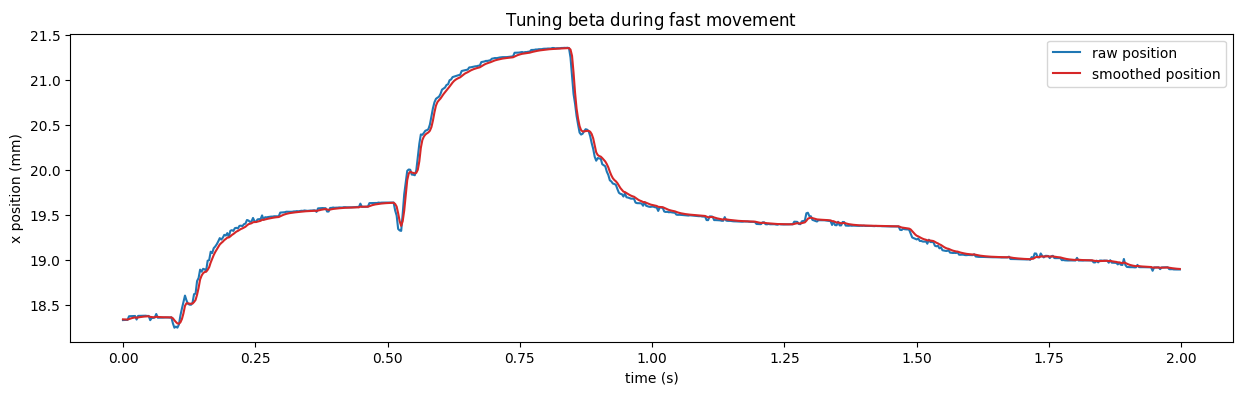
✅ Here we selected beta=1.4
📝 Note
Smoothing the trajectory data is optional for classifying tail bouts. The transformer model was trained on raw tracking data, so it can handle unsmoothed input just as well.