Tail Bout Classification#
In this notebook, we will walk through the process of classifying tail bouts.
Loading dependencies
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.gridspec as gridspec
from megabouts.tracking_data import TrackingConfig, load_example_data, FullTrackingData
from megabouts.config import TailPreprocessingConfig, TrajPreprocessingConfig
from megabouts.preprocessing import TailPreprocessing, TrajPreprocessing
from megabouts.config import TailSegmentationConfig
from megabouts.segmentation import Segmentation, align_traj_array
from megabouts.classification import TailBouts, BoutClassifier
from megabouts.utils import bouts_category_color, bouts_category_name_short
Check if pytorch is running on cpu or gpu:
Show code cell source
import torch
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
print(device)
cpu
Loading tracking data and preprocessing similar to tutorial_Tail_Preprocessing and tutorial_Traj_Preprocessing
df_recording, fps, mm_per_unit = load_example_data("fulltracking_posture")
tracking_cfg = TrackingConfig(fps=fps, tracking="full_tracking")
head_x = df_recording["head_x"].values * mm_per_unit
head_y = df_recording["head_y"].values * mm_per_unit
head_yaw = df_recording["head_angle"].values
tail_angle = df_recording.filter(like="tail_angle").values
tracking_data = FullTrackingData.from_posture(
head_x=head_x, head_y=head_y, head_yaw=head_yaw, tail_angle=tail_angle
)
tail_preprocessing_cfg = TailPreprocessingConfig(fps=tracking_cfg.fps)
tail_df_input = tracking_data.tail_df
tail = TailPreprocessing(tail_preprocessing_cfg).preprocess_tail_df(tail_df_input)
traj_preprocessing_cfg = TrajPreprocessingConfig(fps=tracking_cfg.fps)
traj_df_input = tracking_data.traj_df
traj = TrajPreprocessing(traj_preprocessing_cfg).preprocess_traj_df(traj_df_input)
tail.df.head(5)
| angle | ... | angle_smooth | no_tracking | vigor | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| segments | ... | segments | |||||||||||||||||||
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | ... | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |||
| 0 | -0.101865 | -0.092813 | -0.107645 | -0.110575 | -0.047699 | -0.145887 | -0.130414 | -0.058892 | -0.128705 | NaN | ... | -0.051057 | -0.055168 | -0.055898 | -0.061701 | -0.061091 | -0.069420 | -0.115014 | 0.000183 | True | NaN |
| 1 | -0.082618 | -0.087957 | -0.096951 | -0.092459 | -0.119418 | -0.043354 | -0.099788 | -0.101741 | -0.171555 | NaN | ... | -0.046070 | -0.052906 | -0.056578 | -0.063577 | -0.062286 | -0.064780 | -0.091617 | 0.000321 | True | NaN |
| 2 | -0.093377 | -0.095235 | -0.094292 | -0.105936 | -0.073785 | -0.084193 | -0.144378 | -0.112398 | -0.042585 | NaN | ... | -0.041909 | -0.050905 | -0.056916 | -0.064832 | -0.062943 | -0.060658 | -0.072246 | 0.000430 | True | NaN |
| 3 | -0.092590 | -0.083650 | -0.100938 | -0.088223 | -0.097370 | -0.099559 | -0.101538 | -0.091272 | -0.021459 | NaN | ... | -0.038574 | -0.049166 | -0.056913 | -0.065467 | -0.063063 | -0.057053 | -0.056900 | 0.000512 | True | NaN |
| 4 | -0.086849 | -0.081982 | -0.096705 | -0.118475 | -0.046264 | -0.136459 | -0.115412 | -0.085300 | -0.015487 | NaN | ... | -0.036064 | -0.047688 | -0.056569 | -0.065483 | -0.062646 | -0.053965 | -0.045579 | 0.000565 | True | NaN |
5 rows × 32 columns
Segmentation using tail vigor#
Set the threshold to 20 and apply segmentation to
tail.vigor:
tail_segmentation_cfg = TailSegmentationConfig(fps=tracking_cfg.fps, threshold=20)
segmentation_function = Segmentation.from_config(tail_segmentation_cfg)
segments = segmentation_function.segment(tail.vigor)
We collect the segmented bouts using
extract_tail_arrayandextract_traj_array:
tail_array = segments.extract_tail_array(tail_angle=tail.angle_smooth)
traj_array = segments.extract_traj_array(
head_x=traj.x_smooth, head_y=traj.y_smooth, head_angle=traj.yaw_smooth
)
The arrays contain tail and trajectory data aligned to bout onset:
tail_array: Tail angles for each bout (shape: n_bouts × n_segments × n_timepoints)traj_array: Trajectory data for each bout (shape: n_bouts × 3 × n_timepoints). The 3 dimensions are: x position, y position, and heading angle
print(tail_array.shape, traj_array.shape)
(766, 10, 140) (766, 3, 140)
Optional: Align trajectory data for pre-segmented bouts:
"""
If you're loading pre-segmented data instead of using the segmentation pipeline above,
you'll need to align the trajectory data to the bout onset. This ensures all bouts
are properly oriented relative to their starting position and heading.
"""
# Align trajectory data relative to bout onset
traj_array = align_traj_array(
traj_array=traj_array,
idx_ref=0, # Align to bout onset
bout_duration=tail_segmentation_cfg.bout_duration,
)
Running classifier on the segmented bouts#
Load the classifier, we recommend using
exclude_CS=True. This will avoid assigning bouts to Capture Swim categories if there was no prey during the recording:
classifier = BoutClassifier(tracking_cfg, tail_segmentation_cfg, exclude_CS=True)
Apply the classfier:
classif_results = classifier.run_classification(
tail_array=tail_array, traj_array=traj_array
)
Now we re-segment the bouts using the first half beat as a reference, this improves the alignement of the tail bouts, for this we use the
align_to_onset=Falseoption:
segments.set_HB1(classif_results["first_half_beat"])
tail_array = segments.extract_tail_array(
tail_angle=tail.angle_smooth, align_to_onset=False
)
traj_array = segments.extract_traj_array(
head_x=traj.x_smooth,
head_y=traj.y_smooth,
head_angle=traj.yaw_smooth,
align_to_onset=False,
)
We can use the TailBouts class to store the results of the classifier:
# Format Output:
bouts = TailBouts(
segments=segments,
classif_results=classif_results,
tail_array=tail_array,
traj_array=traj_array,
)
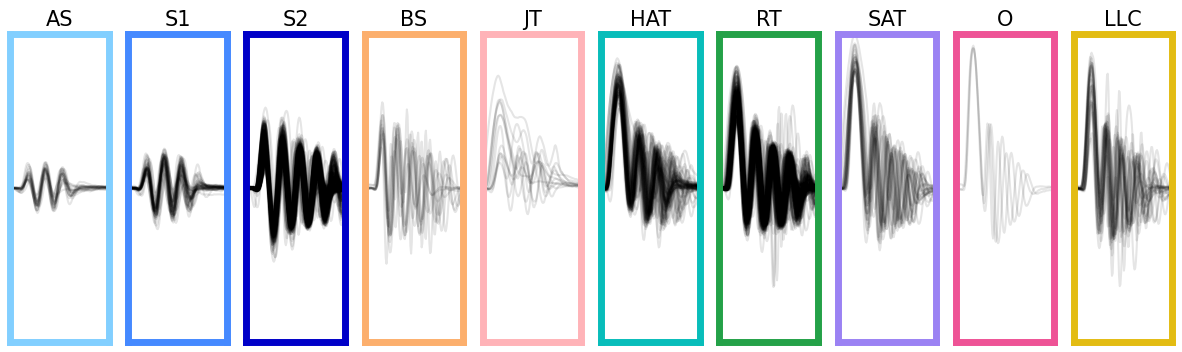
Let’s display the bouts classified, we only display bouts with a classification probability greater than 0.5:
Show code cell source
id_b = np.unique(bouts.df.label.category[bouts.df.label.proba > 0.5]).astype("int")
fig, ax = plt.subplots(facecolor="white", figsize=(15, 4))
ax.spines["top"].set_visible(False)
ax.spines["right"].set_visible(False)
ax.spines["bottom"].set_visible(False)
ax.spines["left"].set_visible(False)
ax.set_xticks([])
ax.set_yticks([])
G = gridspec.GridSpec(1, len(id_b))
ax0 = {}
for i, b in enumerate(id_b):
ax0 = plt.subplot(G[i])
ax0.set_title(bouts_category_name_short[b], fontsize=15)
for i_sg, sg in enumerate([1, -1]):
id = bouts.df[
(bouts.df.label.category == b)
& (bouts.df.label.sign == sg)
& (bouts.df.label.proba > 0.5)
].index
if len(id) > 0:
ax0.plot(sg * bouts.tail[id, 7, :].T, color="k", alpha=0.1)
ax0.set_xlim(0, tail_segmentation_cfg.bout_duration)
ax0.set_ylim(-4, 4)
ax0.set_xticks([])
ax0.set_yticks([])
for sp in ["top", "bottom", "left", "right"]:
ax0.spines[sp].set_color(bouts_category_color[b])
ax0.spines[sp].set_linewidth(5)
plt.show()